Arms-races and the evolution of the Drosophila genome
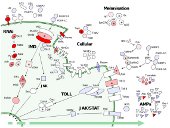 We are interested in understanding how gene function affects the rate of evolution, and particularly whether molecular 'arms-races' might drive a disproportionate number adaptive substitutions.
Because the immune system is thought to coevolve with pathogens (and other parasites), it is expected to evolve rapidly relative to the rest of the genome, and we have used population genetics approaches in Drosophila to look for differences in the rate of adaptive evolution between immunity and non-immunity genes, and between different types of immunity gene.
In Drosophila it is striking that RNAi genes (including those that defend against viruses) show very strong evidence of adaptive evolution, suggesting that they are engaged in an arms race with viruses or parasitic genetic elements.
Our ongoing work focusses on comparative and population genomics of Drosophilidae to understand how selection on the immune system varies among lineages.
We are also working to improve estimates of mutation rate in Drosophila, as a tool to better understand the timescale of selection and evolution, and making experimental attempts to more directly estimate the strength of selection imposed by pathogens on Drosophila.
We are interested in understanding how gene function affects the rate of evolution, and particularly whether molecular 'arms-races' might drive a disproportionate number adaptive substitutions.
Because the immune system is thought to coevolve with pathogens (and other parasites), it is expected to evolve rapidly relative to the rest of the genome, and we have used population genetics approaches in Drosophila to look for differences in the rate of adaptive evolution between immunity and non-immunity genes, and between different types of immunity gene.
In Drosophila it is striking that RNAi genes (including those that defend against viruses) show very strong evidence of adaptive evolution, suggesting that they are engaged in an arms race with viruses or parasitic genetic elements.
Our ongoing work focusses on comparative and population genomics of Drosophilidae to understand how selection on the immune system varies among lineages.
We are also working to improve estimates of mutation rate in Drosophila, as a tool to better understand the timescale of selection and evolution, and making experimental attempts to more directly estimate the strength of selection imposed by pathogens on Drosophila.
The diversity and host range of Drosophila viruses
 Although Drosophila is one of our best models for antiviral immunity in invertebrates, until recently surprisingly little has been known about Drosophila's natural viruses.
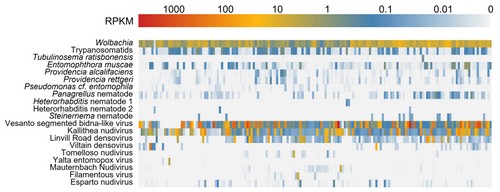
We have used metagenomic sequencing to identify more than 100 new RNA viruses and around 15 DNA virus in wild Drosophila of many different species,and have used RT-PCR surveys and metagenomic approaches to show that many of these viruses are common (and that cell culture infections are almost ubiquitous).
We maintain a comprehensive list of all published Drosophila viruses, and we provide a broad sample of published and unpublished virus and isolate sequences.
Using Drosophila as a model clade, we are using geographic sampling of multiple species to better understand viral host range and patterns of pathogen exchange among hosts.
We have isolated one DNA virus for further study, and we would like to isolate more.
Although Drosophila is one of our best models for antiviral immunity in invertebrates, until recently surprisingly little has been known about Drosophila's natural viruses.
We have used metagenomic sequencing to identify more than 100 new RNA viruses and around 15 DNA virus in wild Drosophila of many different species,and have used RT-PCR surveys and metagenomic approaches to show that many of these viruses are common (and that cell culture infections are almost ubiquitous).
We maintain a comprehensive list of all published Drosophila viruses, and we provide a broad sample of published and unpublished virus and isolate sequences.
Using Drosophila as a model clade, we are using geographic sampling of multiple species to better understand viral host range and patterns of pathogen exchange among hosts.
We have isolated one DNA virus for further study, and we would like to isolate more.
The diversity and host range of Drosophila parasites and pathogens
In parallel with our work on virus discovery, we are using metagenomic approaches to identify and catalogue the distribution and prevalence of common Drosophila pathogens and parasites.
For example, in a pool-seq data of European Drosophila melanogaster (generated by the DrosEU Consortium) we identified four different nematode species (of which three are likely to be parasitic), and large numbers of reads from the fungal pathogen Entomophthora muscae and multiple species of trypanosomatid.
We are continuing this work in collaboration with DrosEU, and in other drosophilid communities.
The evolution of RNAi
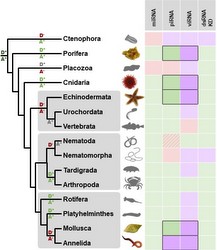 RNA interference has an essential role in mediating defence against viruses and transposable elements. This is well-studied in plants, fungi, nematodes and insects, and so appears to be phylogenetically ancient.
However, in the absence of experimental data from basal eukaryotes (and most animal phyla), the origins and homology between antiviral RNAi mechanisms remains unclear. We are therefore using a metagenomic approach to study viruses and virus-derived siRNAs in a range of non-model lineages.
We also have a broad interest in the way that RNAi pathway genes evolve. This includes both the origins and duplication history of major RNAi gene families, and the role that selection mediated by viruses and/or transposable elements might have played in shaping their sequences.
RNA interference has an essential role in mediating defence against viruses and transposable elements. This is well-studied in plants, fungi, nematodes and insects, and so appears to be phylogenetically ancient.
However, in the absence of experimental data from basal eukaryotes (and most animal phyla), the origins and homology between antiviral RNAi mechanisms remains unclear. We are therefore using a metagenomic approach to study viruses and virus-derived siRNAs in a range of non-model lineages.
We also have a broad interest in the way that RNAi pathway genes evolve. This includes both the origins and duplication history of major RNAi gene families, and the role that selection mediated by viruses and/or transposable elements might have played in shaping their sequences.